





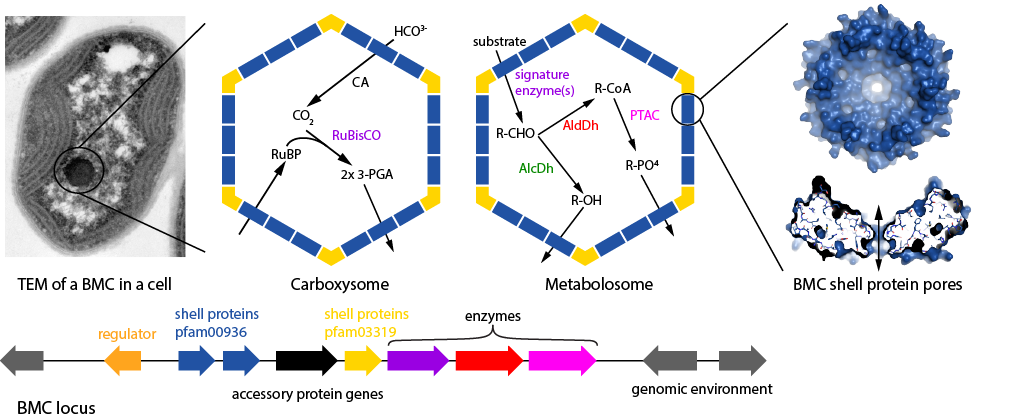
BMC Caller[1] is a tool to analyze sequence data for the presence of Bacterial Microcompartments (BMCs). It also serves as a reference database for BMC types. BMCs are protein-based organelles found in many bacteria[2] that sequester anabolic (Carboxysome) or catabolic (Metabolosome) reactions. The genes encoding the proteins that form the BMCs as well as accessory factors such as regulators or transmembrane transporters for substrates generally cluster as BMC loci. The BMC membrane is composed of unique, conserved protein families. Shell protein oligomers (hexamers, trimers) are typically perforated by pores that selectively allow substrates and products to enter and exit the BMC. BMC Caller has two modes of protein sequence analysis, a locus analysis mode that determines the BMC type, and a shell protein analysis mode that focuses on the proteins forming the shell. The locus database serves as a reference for different BMC types.
This server was established with support from the MSU-DOE Plant Research Laboratory and funding from National Institutes of Health (5R01AI114975-07).